Results
The project was completed in MATLAB using custom functions written to meet the specifications of the project. Results are presented by showing initial images and images that are produced by the algorithm being discussed. In each figure caption image filenames are in parentheses.
The image displayed in this report may be
scaled in the document in order to enhance
readability. However, each image is
available in its entirety if saved from the
web browser. The results presented in this section are given in the same order delineated in the Methods Section.
Part 1
This part
consists of two sections. The first section,
a, studies shape isolation using a
conditional dilation method. The second
part, b, studies the pecstrums of each of
the extracted objects from part a.
Part 1.A
|
|
|
 |
|
Figure
1: A binary image of three shapes. (TheThreeShapes2.gif)
|
Figure
2: A binary images of letters.
(penn256.gif)
|
Figure
2: A binary image of four distinct objects.
(match1.gif) |
A function was created to do the object
extraction. It has two parameters: the
number of objects and the binary image
containing the objects. The algorithm works
by first scanning the entire image until it
hits an 'ON' pixel (level 1 if normalized).
Once such a pixel was found, that pixel was
dilated with a structuring element B. The
resulting dilation was intersected with the
original image and then stored in a matrix
V. Upon each iteration, V was checked
to see if it changed from the previous value
of V. If V did not change, the object
was found and the algorithm stopped
iterating. Thus, upon V reaching a steady
state, the iterations stop.
B = 111
111
111
Next, the bounding box for the object
recently found was computed. This was done
by scanning the image V and determining the
greatest and smallest values at which a ON
pixel existed for both the X and Y axis.
It should be noted that the bounding box was
constructed to INCLUDE the box itself as
part of the image. Thus, the values computed
for the top, bottom, left, and right of the
object can be solely used without
modification to quickly extract the object.
The box was drawn around the object using a
for loop and is the same color as the image.
The images were colored such that they
would be easily resolved as unique shades of
gray to the human eye. This is why the
number of objects was passed to the
findObjects function. This enabled us to
calculate the color values of each object
before running the iterative algorithms
described above. The found objects in their
image were written out to disk with the
appropriate color for identification.
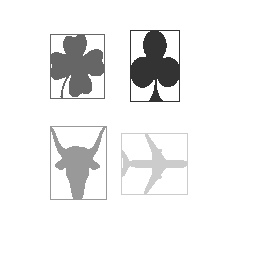
The results of the object identification
are shown below (Figures 4 - 6). Note that
each object is a different shade of gray.
The background of the image each of these
objects are contained in is white
|
|
|
 |
|
Figure
4: Three
shapes
extracted. (TheThreeShapes2_objects.png)
|
Figure
5: Nine
letters
extracted
(penn256_objects.png)
|
Figure
6: Four distinct
objects extracted.
(match1_objects.gif) |
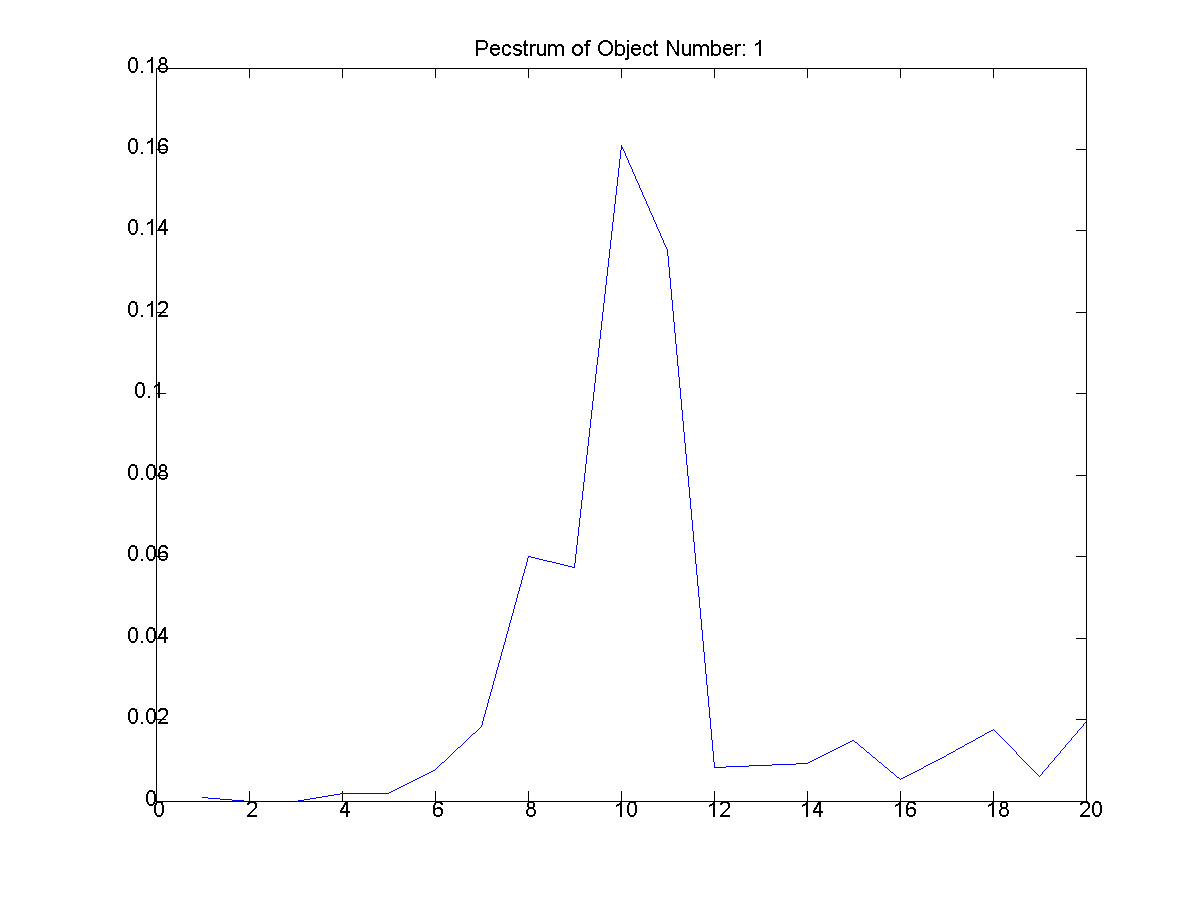
Upon completion of the bounding box,
the pecstrum of each object found in the
input image was computed. The following
algorithm was used to compute the pecstrum.
B = 111
111
111
pecstrum = f(n) = [ m(XnB)
- m(X(n+1)B) ] / [m(X)]
where m is the 'area' of the object.
The results pecstrum of each object is shown
below (Figures 7-22). Note that the
domain of the pecstrum is discrete and not
continuous as depicted by the MATLAB PLOT
command. However, the values at each point
in the discrete domain are correct.
Each pecstrum graph can be blown up by
clicking on it.
From Figure 1, these three shapes were
extracted. The first object filename is
provided as the other are of similar
pattern.
 |
 |
 |
 |
 |
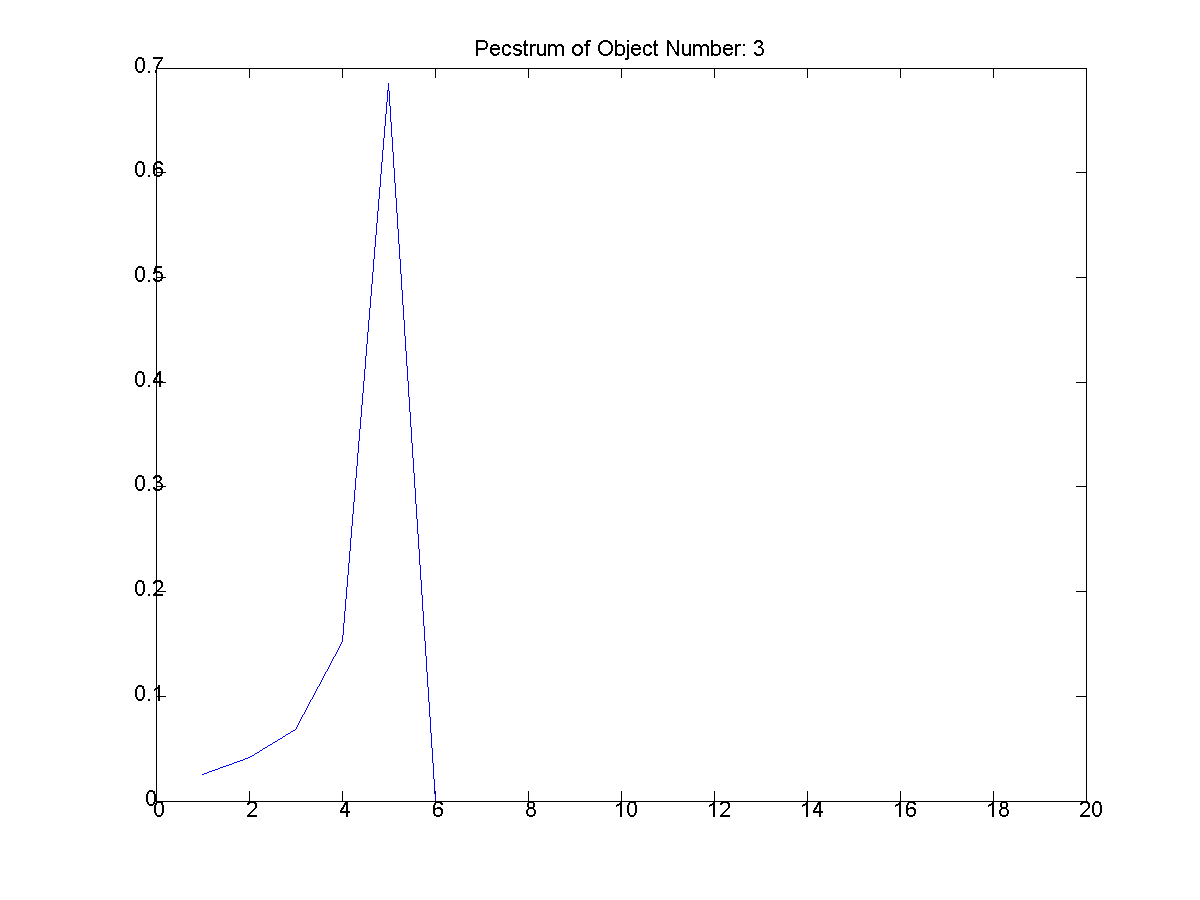 |
| Figure
7: The first extracted object and
its pecstrum below it. (T: thethreeshapes2/object_1.png,
B:thethreeshapes2/object_1_pectsrum.png) |
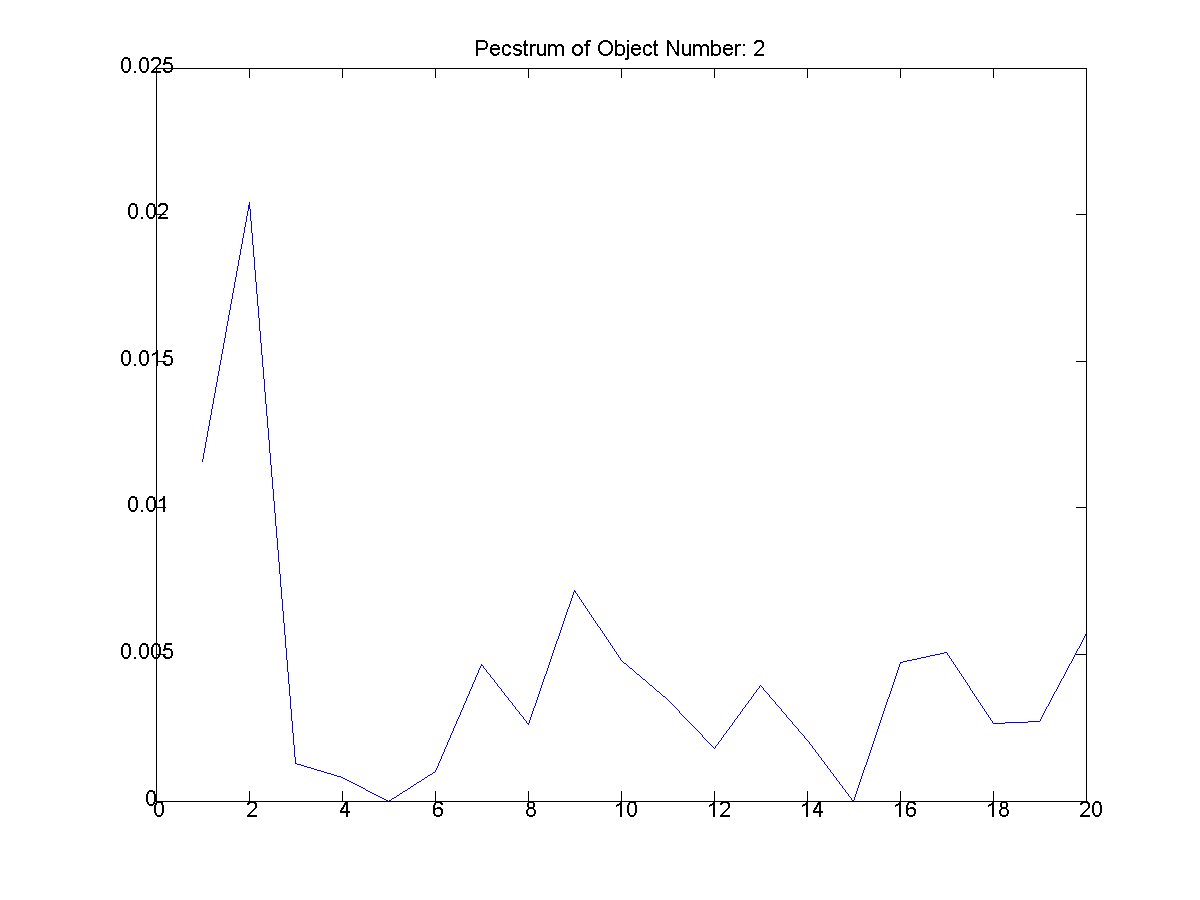
Figure
8: The second extracted object and
its pecstrum below it. |
Figure
9 The third extracted object and
its pecstrum below it. |
From Figure 2, these three shapes were
extracted. The table below represents Figures
10 -18. Each extracted objects
pecstrum is adjacent to itself. The
first object filename is provided as the
other are of similar pattern.
From Figure 3, these four shapes were
extracted. The first object filename is
provided as the other are of similar
pattern.
Figure 19: The
first extracted object and its
pecstrum adjacent to it.
(L: match1/object_1.png, match1/object_1_pectsrum.png) |
 |
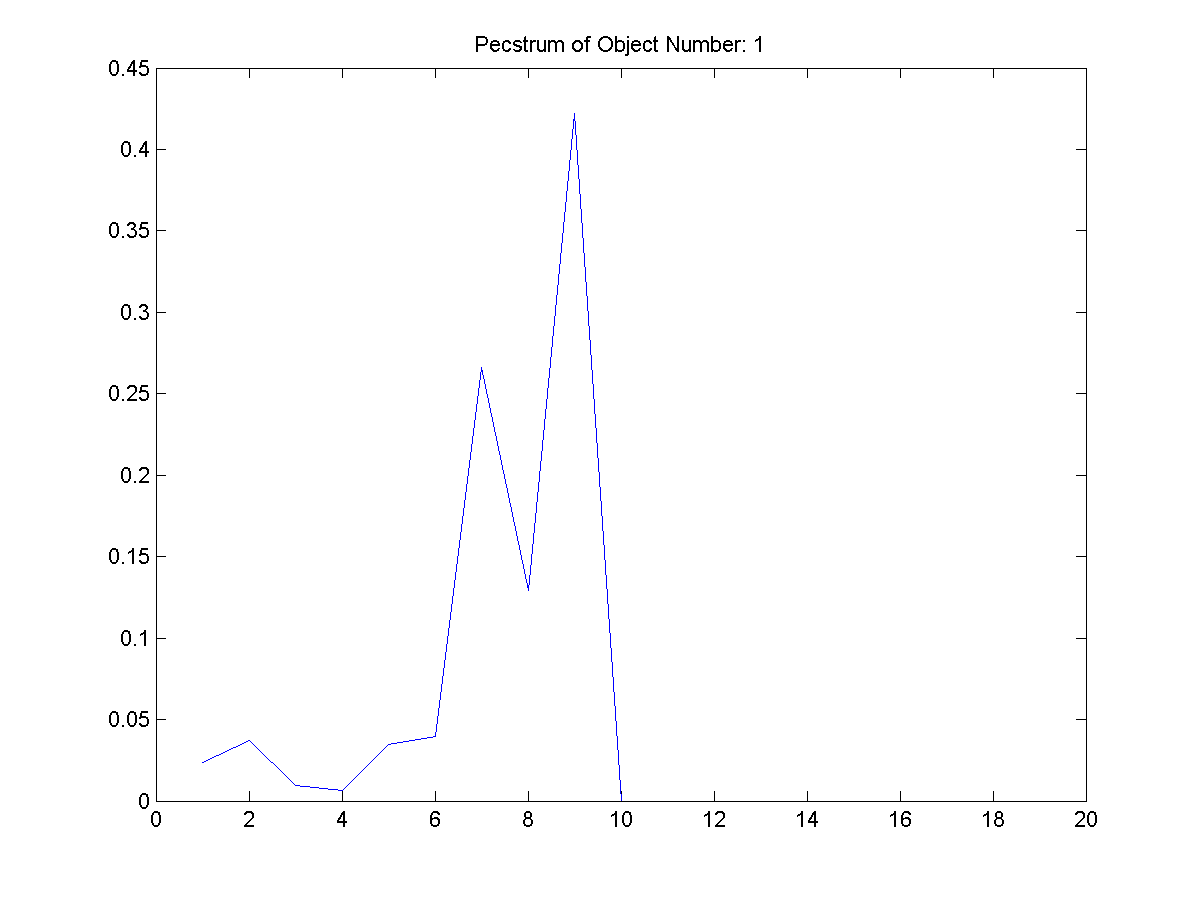 |
| Figure 20: The
second extracted object and its
pecstrum adjacent to it. |
 |
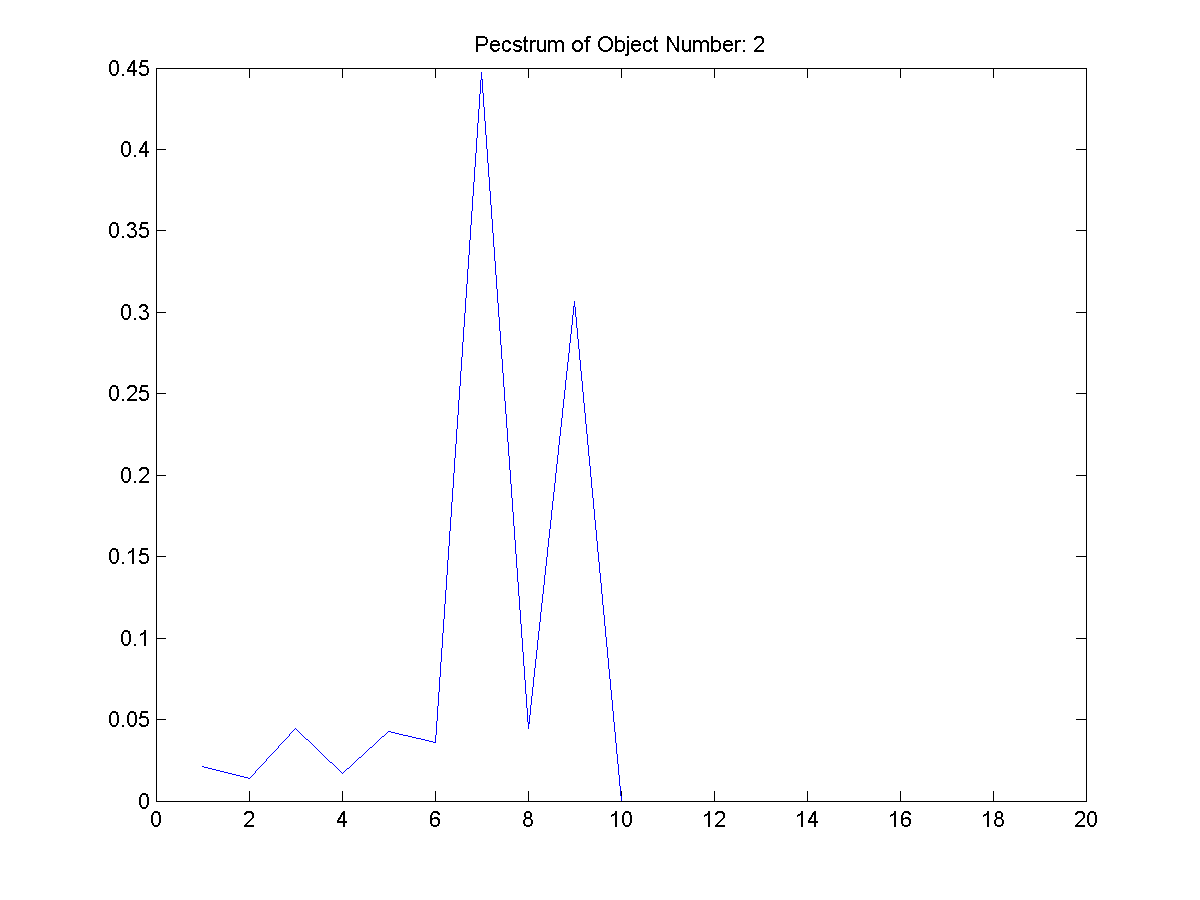 |
| Figure 21: The
third extracted object and its
pecstrum adjacent to it. |
 |
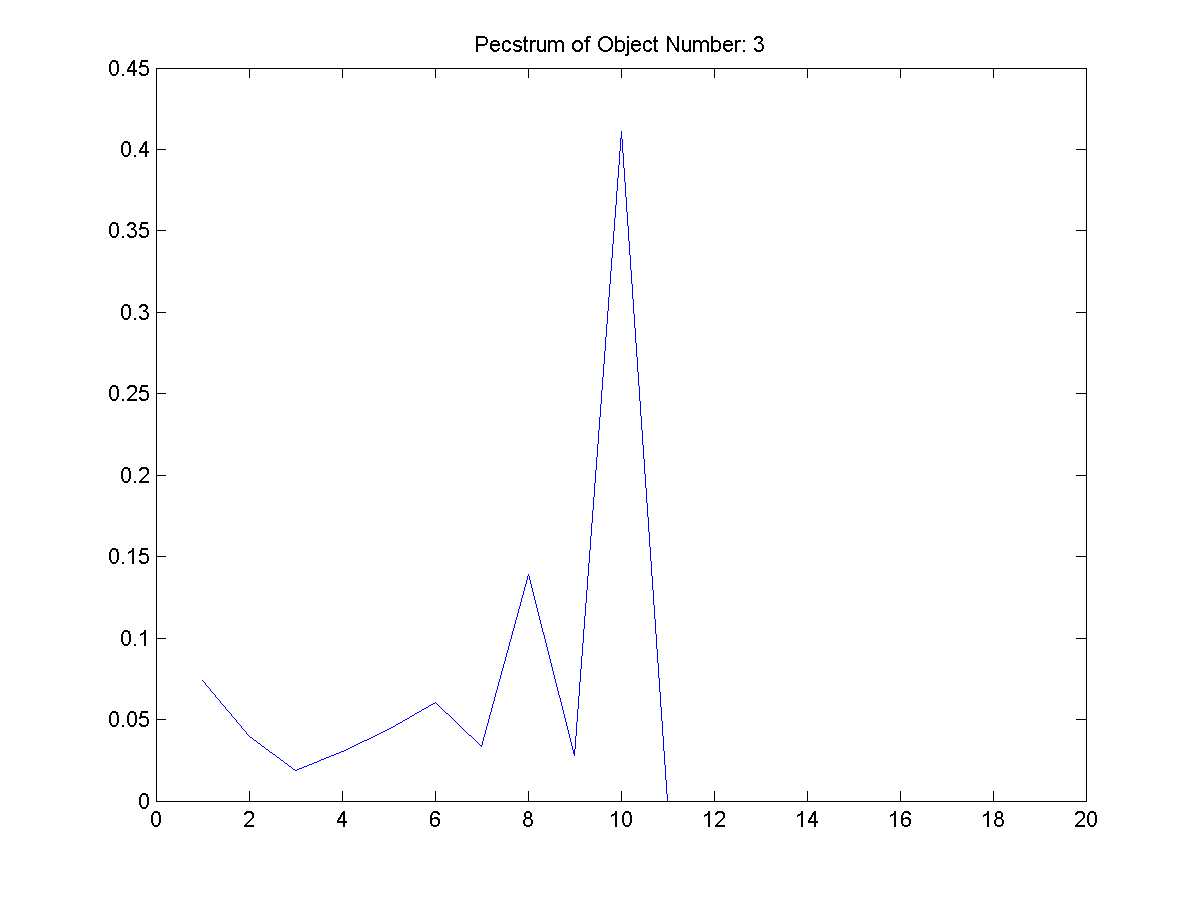 |
| Figure 22: The
fourth extracted object and its
pecstrum adjacent to it. |
 |
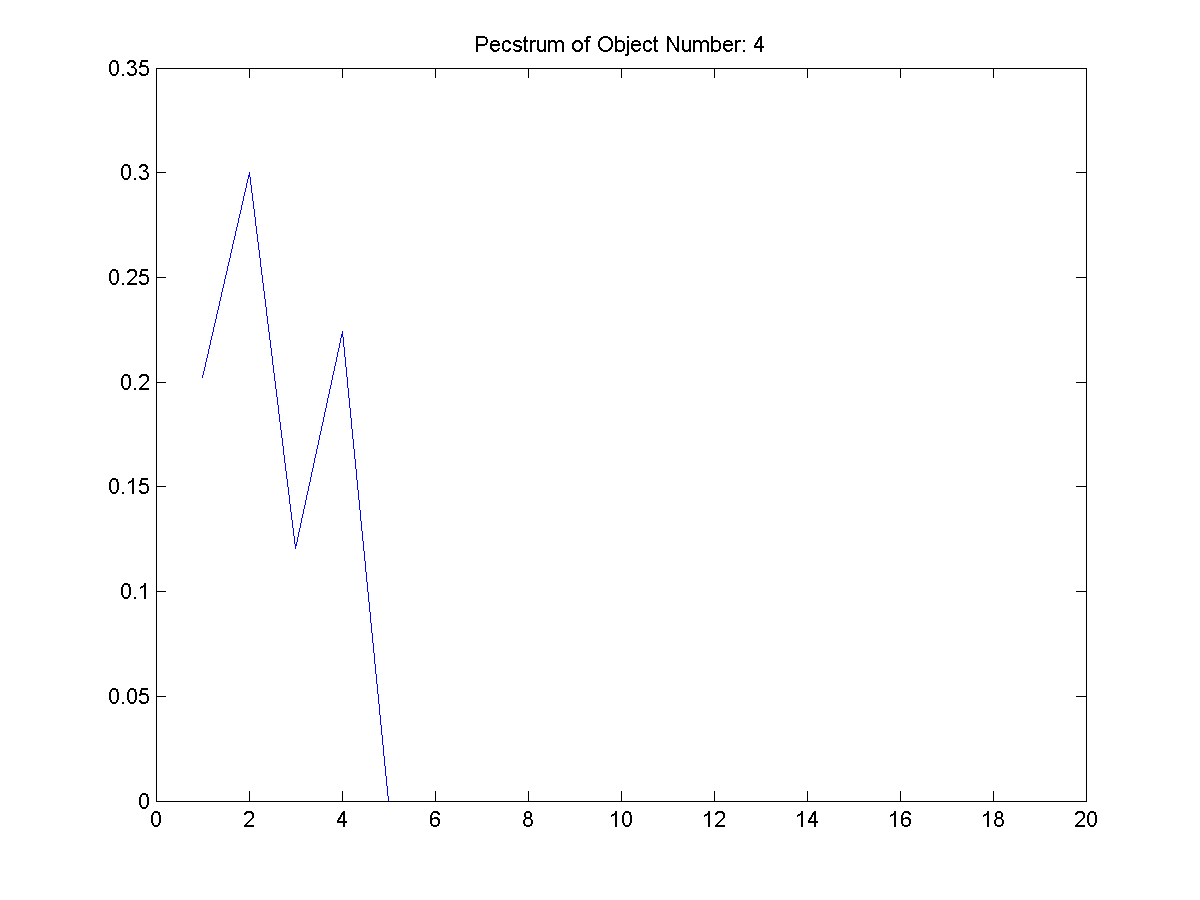 |
Part 1.B
The second section of Part 1, section
b, was to compute the pecstrums as
shown above. Figure 8, from its
pecstrum, appears to be the most complex
object extracted from the three
images. Its pecstrum has many delta
functions implying the object is very
"jaggy". Typically jaggy objects
are not have little repetition (patterns)
and symmetry and can be described as complex
because so. Many of the other objects in the
data set have straight edges or have
symmetric properties. Thus, they are few
deltas in their pecstrum.
Pecstrum is somewhat analogous to
frequency spectrum. It should be noted that
similar to a frequency spectrum, the sum of
the area under each pecstrum function is
equal to one.
Part 2
This part of the project focused on morphological
skeletonization. The process of skeletonization creates a set of skeletons for a given image. Each skeleton represents a degree of detail in the original image. The skeletons are generated sequentially, with the first skeletongs Sk(0), Sk(1), etc. containing the smallest details in the original image. The larger order skeletons
Sk(n), Sk(n-1), etc. contain the "meat" of the objects in the original image. There are a variety of variables in
skeletonization, including the way in which it is applied and the structuring element used to apply it. Generally, one would like to use a circular structuring element of relatively small radius with respect to the object. In the
discrete domain, however, the circle must be approximated and cannot be smaller that 5x5. This structuring element, shown as "B" below, was chosen for this part of the project, as recommended by Pitas and
Venetsanopoulos.
B = 0 1 1 1 0
1 1 1 1 1
1 1 1 1 1
1 1 1 1 1
0 1 1 1 0
Part 2.A
The goal of this part is to implement and test the skeletonization algorithm on two test images. The images chosen are show in in figures
23 and 25. Results for each are in Figures
24 and 24 The algorithm implemented is document in P&V 6.8.7-6.8.10 and Figure 6.8.4 (a). The output of this routine is a large matrix of skeletons, Si(X), and the union of these. The latter output constitutes the morphological skeleton of the original image.
| Figure
23: Original image (match1.gif) |
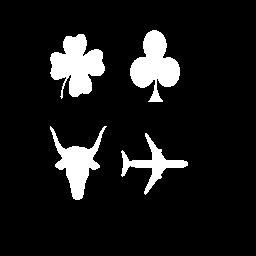 |
| Figure
24: Skeleton of match1.gif (match1skel.png) |
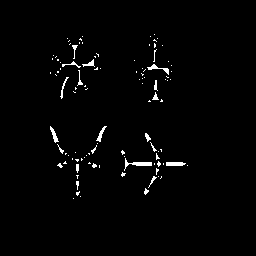 |
| Figure
25:Original image
(bear.gif) |
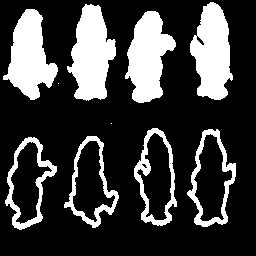 |
| Figure
26: Skeleton of bear.gif
(bearskel.png) |
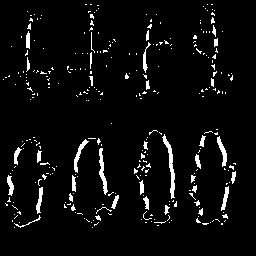 |
Comparing results to P&V Figure 6.8.4 it is clear that the algorithm performs as desired. Particularly remarkable effects are the generally irregular shape of the skeletons and preservation of the salt noise pixels in the bear image. The morphological skeletonization routine is clearly not perfect, but it achieves the desired result of finding the medial axis of objects. A subsequent operation that performs object connection and removes outliers would enhance the usefulness of the result.
Part 2.B
The goal of this part is to take the skeletons (S0(X), S1(X1), ..., SN(X)) from Part 2.A and attempt to partially reconstruct the original image. The reconstruction will be based on the "meat" of the image, that is SN(X), SN-1(X), SN-2(X) and SN-3(X). The alorithm used is documented in P&V 6.8.10 and Figure 6.8.5 (b). The routine that accomplishes the partial reconstruction accepts a large matrix of all skeletons, Si(X), and picks of the four largest. These are then feed through the algorithm. For comparision, a full reconstruction of bear.gif is provided to demonstrate the effectiveness of the reconstruction algorithm.
| Figure 27: Partial reconstruction of the match1.gif skeleton
(matchrecon.png) |
 |
| Figure
28: Set difference between skeleton of match1.gif and partial reconstruction
(matchrecon_diff.png) |
 |
| Figure 29: Partial reconstruction of the bear.gif skeleton
(bearrecon.png) |
 |
| Figure 30
Set difference between skeleton of bear.gif and partial reconstruction
(bearrecon_diff.png) |
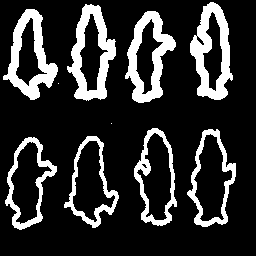 |
| Figure 31: Full reconstruction of the bear.gif skeleton
(bearrecon_FULL.png) |
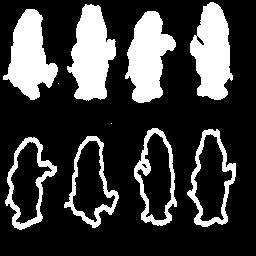 |
| Figure
32: Set difference between skeleton of bear.gif and full reconstruction
(bearrecon_FULL_diff.png) |
 |
It is clear from the results in Figures
27 and 29 that the partial reconstruction using only the four highest order skeletal components removes details. This is clearly realized when examining the partial reconstruction resutls for bear.gif and realizing that the "outline bears" are completely removed. Since the outlines are relatively thin, they are not present in the four highest order skeletal components. Note that if the outlines had been the only thing present in the image (the solid bears were absent) they would be present in the partial reconstruction as the highest order skeletal components would not be as high order as those requried to skeletonize the solid bears. Finally, observing the results in Figures
31 and 32 one can see that the skeletonization algorithm allows perfect reconstruction of the original image if all skeletal components are used (this is shown because the set difference between the full reconstruction and the original image is zero).
Part 2.C
All results and figures for Part 2 are given and discussed in their respective sections (A or B) above.
Summary
The results presented here are relatively straight forward. All outcomes achieved were as expected in each experiment, as we were able to predict what we should see and explain each result.
Practical uses and analysis of basic morphological operations
and algorithms are demonstrated.
|